Attribution Priors
A repository for training explainable models using attribution priors.
This repository contains tools for connecting the machine learning topics of model priors and model explanations with a new method called attribution priors, discussed in our paper "Learning Explainable Models Using Attribution Priors". This package contains:
- A differentiable axiomatic feature attribution method called expected gradients.
- Tensorflow and PyTorch operations to directly regularize expected gradients attributions during training.
- Examples of how arbitrary differentiable functions of expected gradient attributions can be regularized during training to encode prior knowledge about a modeling task.
For more guidance about how to use this repository/how to train with attribution priors, one of the quickest demos for PyTorch is available in the
Convergence Demo.ipynb notebook. For quick demos in Tensorflow, see the example_usage.ipynb notebook
(for older versions of TensorFlow) and the example_usage_tf2.ipynb notebook (for TensorFlow 2.0 and above)
in the top level directory of this repository, and the Installation and Usage Section of this README.
Compatability
The code in this repository was written to support TensorFlow versions r1.8 and up, and works with both Python 2 and 3. If you are using TensorFlow with eager execution/TensorFlow 2.0 and above, see Training with Eager Execution. If you are training with TensorFlow Sessions (old-school TensorFlow), see Training with TensorFlow Sessions. We also now have support for PyTorch, which has been tested with Python 3 - see Training with PyTorch.
Code has been tested for GPU compatability on a Lambda Blade GPU Server running CentOS 7.8, and for CPU compatibility on a MacBook Pro running macOS Catalina 10.15.4.
Installation
The easiest way to install this package is by cloning the repository:
git clone https://github.com/suinleelab/attributionpriors.git
Installation should take less than a minute.
Demo
The quickest demo to run the code is the Convergence Demo.ipynb notebook, available in the main directory. This notebook should run in a matter of minutes using only cpu, and also illustrates the benefits of expected gradients feature attributions over other feature attribution methods for attribution priors. For quick demos in Tensorflow, see the example_usage.ipynb notebook
(for older versions of TensorFlow) and the example_usage_tf2.ipynb notebook (for TensorFlow 2.0 and above)
in the top level directory of this repository, and the Installation and Usage Section of this README.
Examples
So what exactly are attribution priors and why would you want to use them? The examples here provide three ways in which you can use attribution priors to improve network performance and interpretability. We use these examples in our paper. However, attribution priors are not limited to the examples here.
Image Data (mnist)
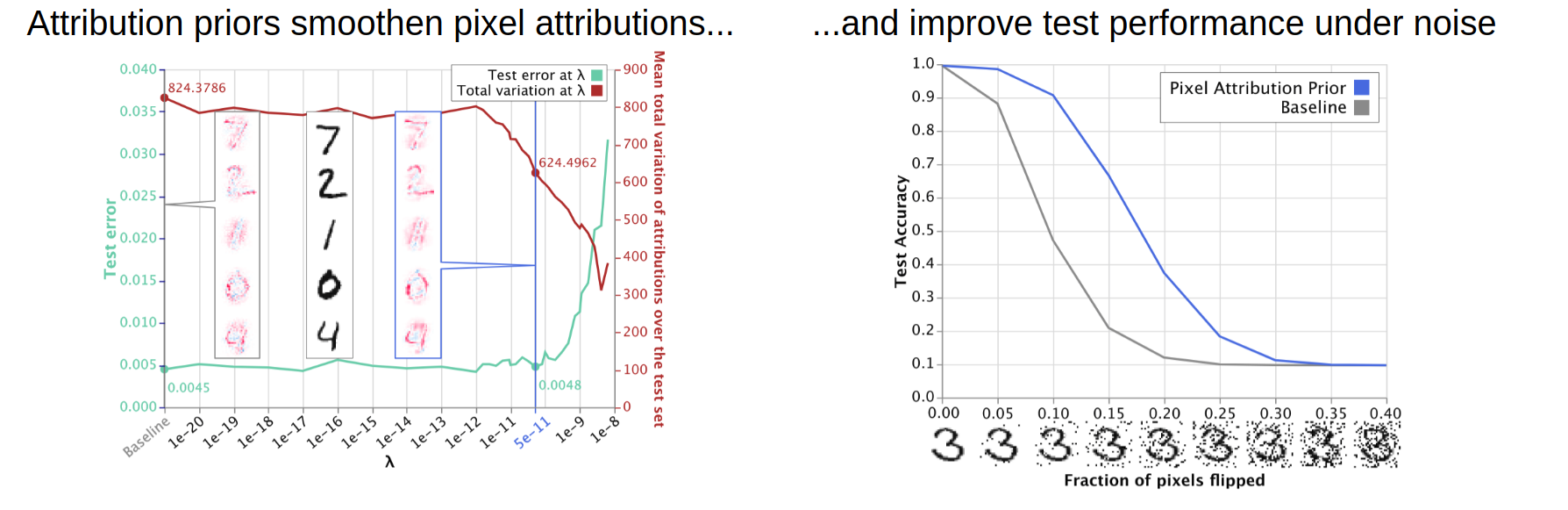
In the mnist folder, we give examples about how to train models that have smoother attributions over pixels, which in turn
leads to better performance on noisy test data. Click through the notebooks in that folder to see more.
Gene Expression Data (graph)
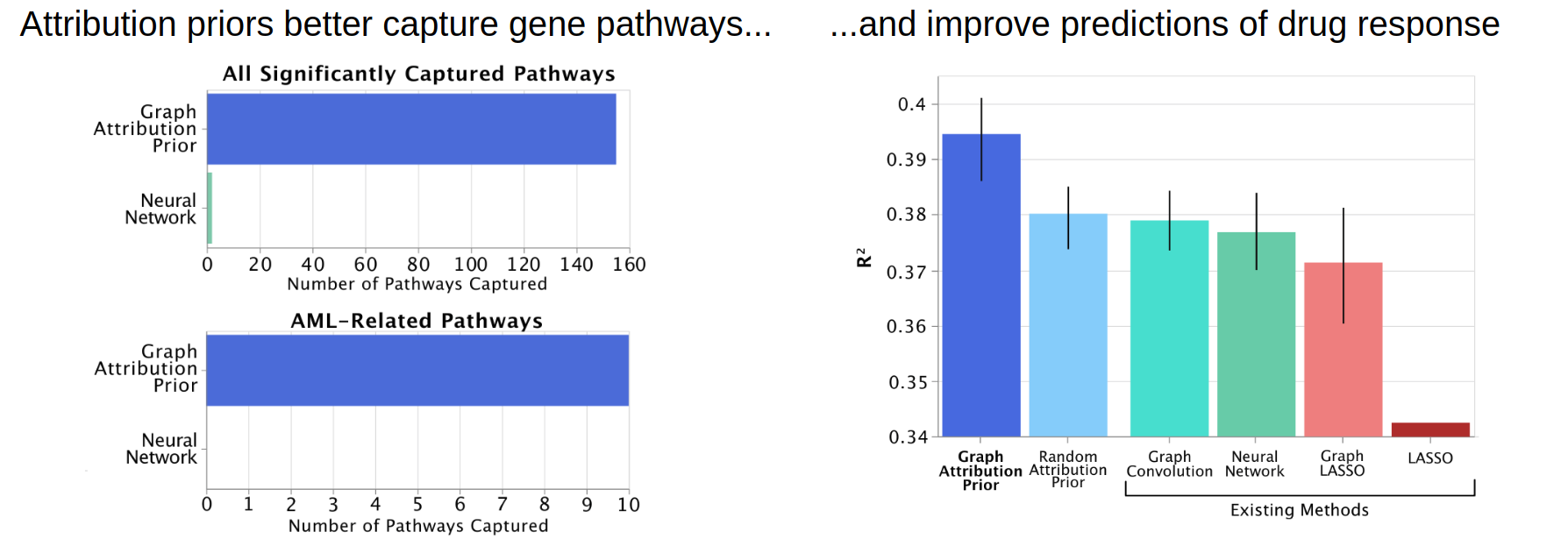
In the graph folder, the notebook shows how penalizing differences between the attributions of neighbors in an arbitrary graph
connecting the features can be used to incorporate prior biological knowledge about the relationships between genes,
yield more biologically plausible explanations of drug response predictions, and improve test error.
Tabular Data (sparsity)
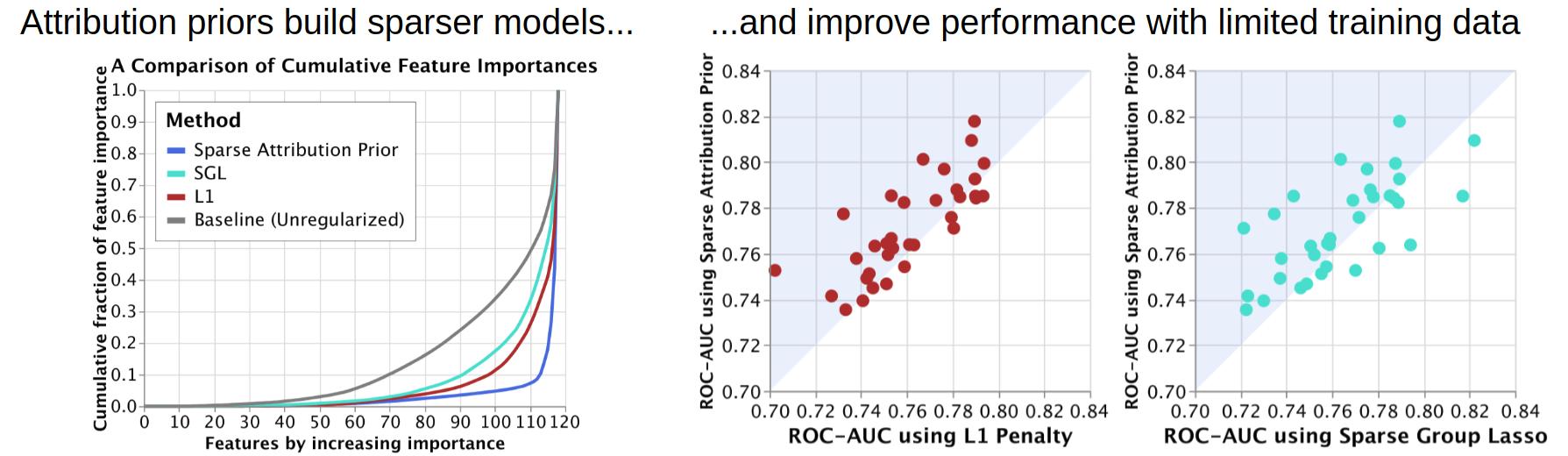
In the sparsity folder, the notebook shows how encouraging inequality in the distribution of feature attributions
can build sparser models that can perform more accurately when training data is limited.
Usage: Training with Eager Execution
This code provides an API for users who are using TensorFlow with eager execution, which is the default in TensorFlow 2.0 and above. The API change is rather simple in eager exceution and follows the following steps:
1: Importing
#Other import statements...
from attributionpriors import eager_ops2: Manually writing the train_step
Where normally you would write code like this:
@tf.function
def train_step(inputs, labels, model):
with tf.GradientTape() as tape:
tape.watch(inputs)
predictions = model(inputs, training=True)
pred_loss = loss_fn(labels, predictions)
total_loss = pred_loss
if len(model.losses) > 0:
regularization_loss = tf.math.add_n(model.losses)
total_loss = total_loss + regularization_loss
gradients = tape.gradient(total_loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))Now you should add the following lines:
@tf.function
def train_step(inputs, labels, model):
with tf.GradientTape() as tape:
tape.watch(inputs)
predictions = model(inputs, training=True)
pred_loss = loss_fn(labels, predictions)
total_loss = pred_loss
if len(model.losses) > 0:
regularization_loss = tf.math.add_n(model.losses)
total_loss = total_loss + regularization_loss
+ attributions = eager_ops.expected_gradients(inputs, labels, model)
+ attribution_loss = ap_loss_func(attributions, model)
+ total_loss = total_loss + lamb * attribution_loss
gradients = tape.gradient(total_loss, model.trainable_variables)
optimizer.apply_gradients(zip(gradients, model.trainable_variables))where ap_loss_func is some loss function on top of your attributions, and lamb is a scalar
penalty controlling the trade-off between penalizing attributions and your standard training loss.
In the image case, we use tf.reduce_mean(tf.image.total_variation()), the mean of the total variation across the attributions.
And that's it! If you want a more in-depth example, see the example_usage_tf2.ipynb notebook.
Why can't we use the fit function?
Overloading the fit function is, well, more difficult and makes it harder to specify complex penalties on your attributions. It would require sub-classing the tf.keras.model API, which requires handling a variety of edge cases. If you want to take on this project, feel free to do so, but we don't have plans to support it as of right now.
Usage: Training with TensorFlow Sessions
This package provides a simple API that can be used to define attribution priors over neural networks that can be dropped in to existing TensorFlow code. In order to train using our implementation of attribution priors in your own code, you need to follow the following four steps:
1: Importing
#Other import statements...
from attributionpriors.ops import AttributionPriorExplainer2: Wrapping your input tensor using an AttributionPriorExplainer object
Where normally you would write code like this:
input_op = get_input() #Returns some tensor of shape [batch_size, feature_dim0, feature_dim1, ...] that you feed into a TensorFlow modelAdd the following lines in green:
+ explainer = AttributionPriorExplainer()
input_op = get_input()
+ input_op, train_eg = explainer.input_to_samples_delta(input_op)input_op can be a placeholder, a constant, or any other TensorFlow tensor that you use to feed data, like images or feature vectors, into a model.
The above code modifications create operations that alternate between feeding your model normal input and input interpolated between a sample and a reference.
This is used to compute expected gradients.
3: Defining the Expected Gradients Tensor
The code for getting your expected gradients tensor depends on whether your model outputs a single number (e.g. in regression), or multiple (e.g. in classification tasks).
Single-output Tasks (like regression)
If your code to define your model normally looks like this:
output_op = model(input_op) #output_op is a [batch_size] shaped tensor of floatsAdd the following line in green:
output_op = model(input_op)
+ expected_grads_op = explainer.shap_value_op(y_pred, cond_input_op)Multi-output Tasks (like classification)
If instead, you are predicting multiple classes, you need to define which class you want to take expected gradients with respect to. Although our repository supports taking expected gradients with respect to ALL output classes (simply by using the code above in the single-output tasks), it is more computationally efficient and more intuitive to take expected gradients with respect to the true class. That is, if you have an image of, say, a car, you should take attributions with respect to the car output class. Our repository supports this as follows:
label_op = get_labels() #label_op should be a [batch_size] shaped tensor of integers specifying which class each input in input_op belongs to
output_op = model(input_op) #output_op is a [batch_size] shaped tensor of floats
+ expected_grads_op = explainer.shap_value_op(y_pred, cond_input_op, label_op)This will return a tensor operation that represents attributions with respect to the true class of each example.
4: Training with an Attribution Prior
First, you need to define a loss function with respect to the expected_grads_op tensor. You can look to the examples section of the README for some examples
of attribution priors, or you can read our original paper. For example, we use the total variation loss for images to get models with smoother explanations:
eg_loss_op = tf.reduce_mean(tf.image.total_variation(expected_grads_op))
eg_train_op = tf.train.AdamOptimizer(learning_rate).minimize(eg_loss_op)Once you have an operation that trains your eg loss, you can call it alternating with your normal loss. If your code normally looks like this:
for i in range(num_training_steps):
sess.run(train_op)Instead you should write:
for i in range(num_training_steps):
- sess.run(train_op)
+ batch_input, batch_labels, _ = sess.run([input_op, label_op, train_op])
+ sess.run(eg_train_op, feed_dict={train_eg: True,
+ input_op: batch_input,
+ label_op: batch_labels})It is important that you set train_eg: True when you are running any operation related to the expected gradients tensor.
If you do not do so, the code will give strange errors.
And that is all there is to it! Your code will simultaneously minimize both your task objective and also whatever attribution prior you defined.
Usage: Training with PyTorch
This code provides an API for users who are using PyTorch to train their models.
1: Importing
#Other import statements...
from attributionpriors.pytorch_ops import AttributionPriorExplainer2: Initializing AttributionPriorExplainer
Before training, initialize the AttributionPriorExplainer object with the PyTorch Dataset object you want to use as background (we recommend using the full training dataset), the batch size, and the k parameter (number of background references per foreground samples).
APExp = AttributionPriorExplainer(background_dataset, batch_size,k=1)3: Adding Expected Gradients Calculation to Training Step
Where your normal training loop in PyTorch might look like the following...
for features, labels in train_loader:
features, labels = features.cuda().float(), labels.cuda().float()
optimizer.zero_grad()
outputs = model(features)
loss = torch.nn.MSELoss(outputs, labels)
loss.backward(retain_graph=True)
optimizer.step()
train_losses.append(loss.item())Now simply add the following lines to, for example, add an L1 penalty on the expected gradients...
for features, labels in train_loader:
features, labels = features.cuda().float(), labels.cuda().float()
optimizer.zero_grad()
outputs = model(features)
- loss = torch.nn.MSELoss(outputs, labels)
+ expected_gradients = APExp.shap_values(model,features)
+ attribution_prior = torch.norm(expected_gradients, p=1)
+ loss = torch.nn.MSELoss(outputs, labels) + attribution_prior
loss.backward(retain_graph=True)
optimizer.step()
train_losses.append(loss.item())