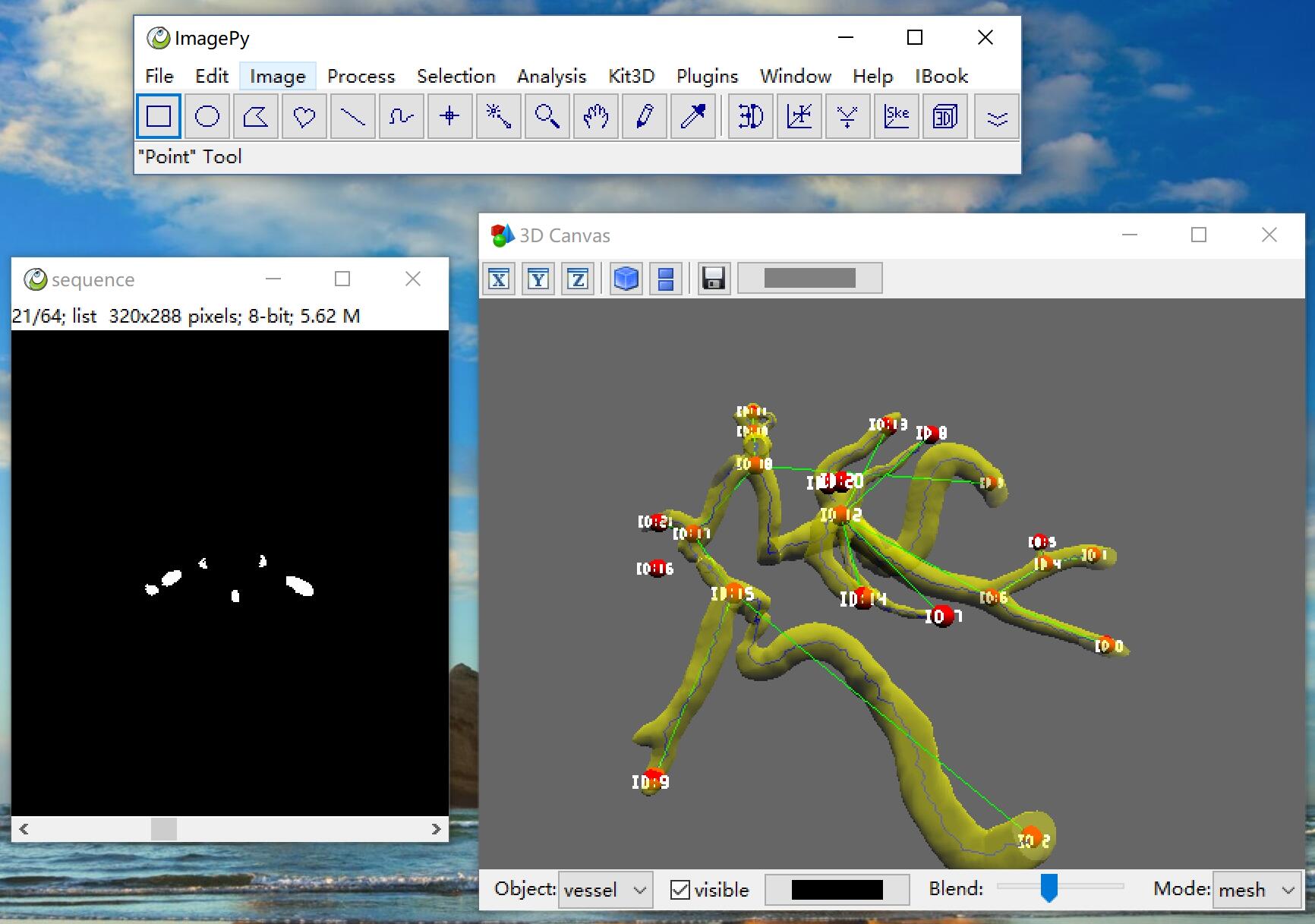
Skeleton Network
build net work from nd skeleton image
graph = sknw.build_sknw(ske, multi=False, iso=True, ring=True, full=True)
ske: should be a nd skeleton image
multi: if True,a multigraph is retured, which allows more than one edge between two nodes and self-self edge.
iso: if True, return one-pixel node
ring: if True, return ring without any branch (and insert a self-connected node in the ring)
full: if True, every edge start from the node's centroid. else touch the node block but not from the centroid.
return: is a networkx Graph object
graph detail:
graph.nodes[id]['pts'] : Numpy(x, n), coordinates of nodes points
graph.nodes[id]['o']: Numpy(n), centroid of the node
graph.edge(id1, id2)['pts']: Numpy(x, n), sequence of the edge point
graph.edge(id1, id2)['weight']: float, length of this edge
if it's a multigraph, you must add a index after two node id to get the edge, like: graph.edge(id1, id2)[0].
Build Graph:
build Graph by Skeleton, then plot as a vector Graph in matplotlib.
from skimage.morphology import skeletonize
from skimage import data
import sknw
# open and skeletonize
img = data.horse()
ske = skeletonize(~img).astype(np.uint16)
# build graph from skeleton
graph = sknw.build_sknw(ske)
# draw image
plt.imshow(img, cmap='gray')
# draw edges by pts
for (s,e) in graph.edges():
ps = graph[s][e]['pts']
plt.plot(ps[:,1], ps[:,0], 'green')
# draw node by o
nodes = graph.nodes()
ps = np.array([nodes[i]['o'] for i in nodes])
plt.plot(ps[:,1], ps[:,0], 'r.')
# title and show
plt.title('Build Graph')
plt.show()Find Path
then you can use networkx do what you want
3D Skeleton
sknw can works on nd image, this is a 3d demo by mayavi
About ImagePy
https://github.com/Image-Py/imagepy
ImagePy is my opensource image processihng framework. It is the ImageJ of Python, you can wrap any numpy based function esaily. And sknw is a sub module of ImagePy. You can use sknw without any code.